Searching annotations
Bystro supports multi-term natural-language search queries for all annotation fields
To start, simply type annotation terms into the search bar
Try searching terms that interest you, such as a gene name, site type or mutation type:
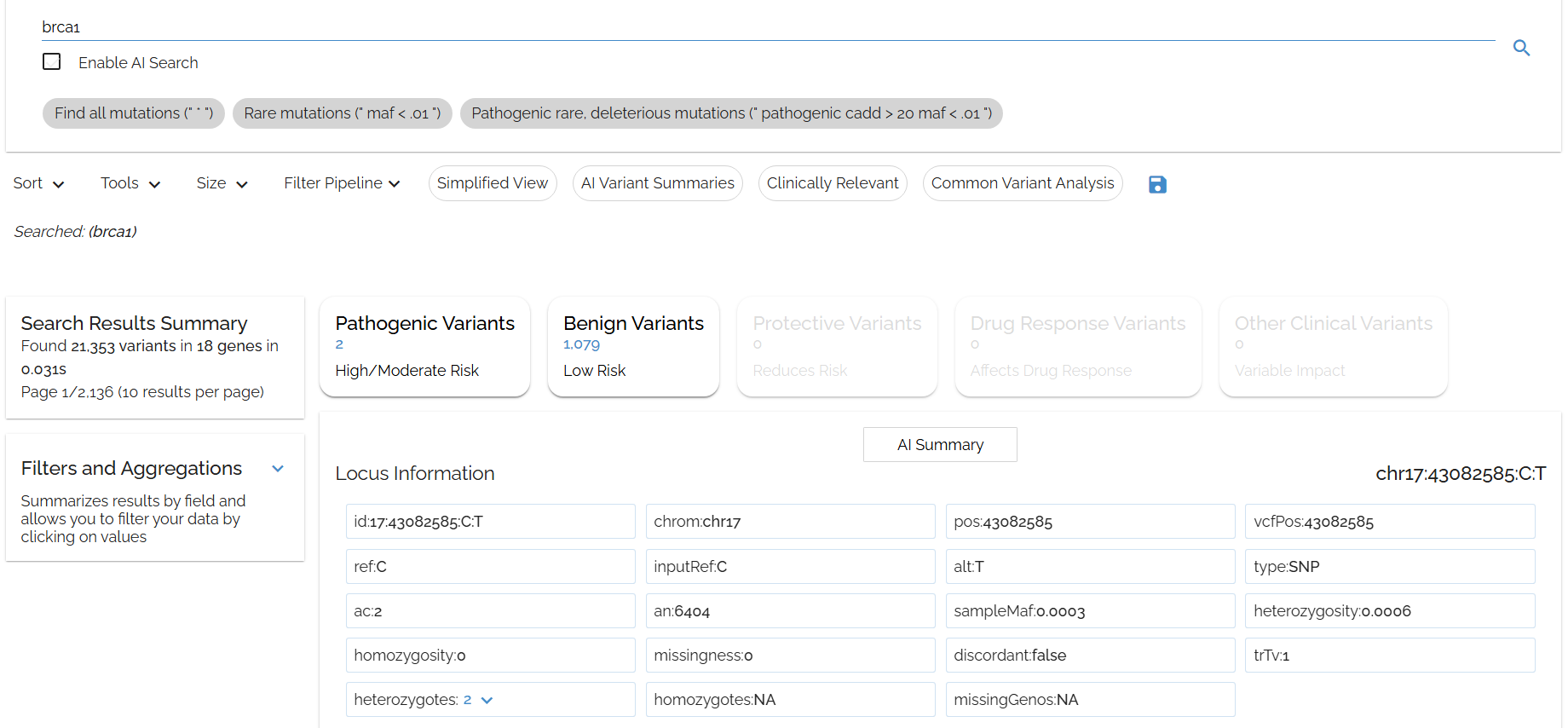
Typing in: brca1
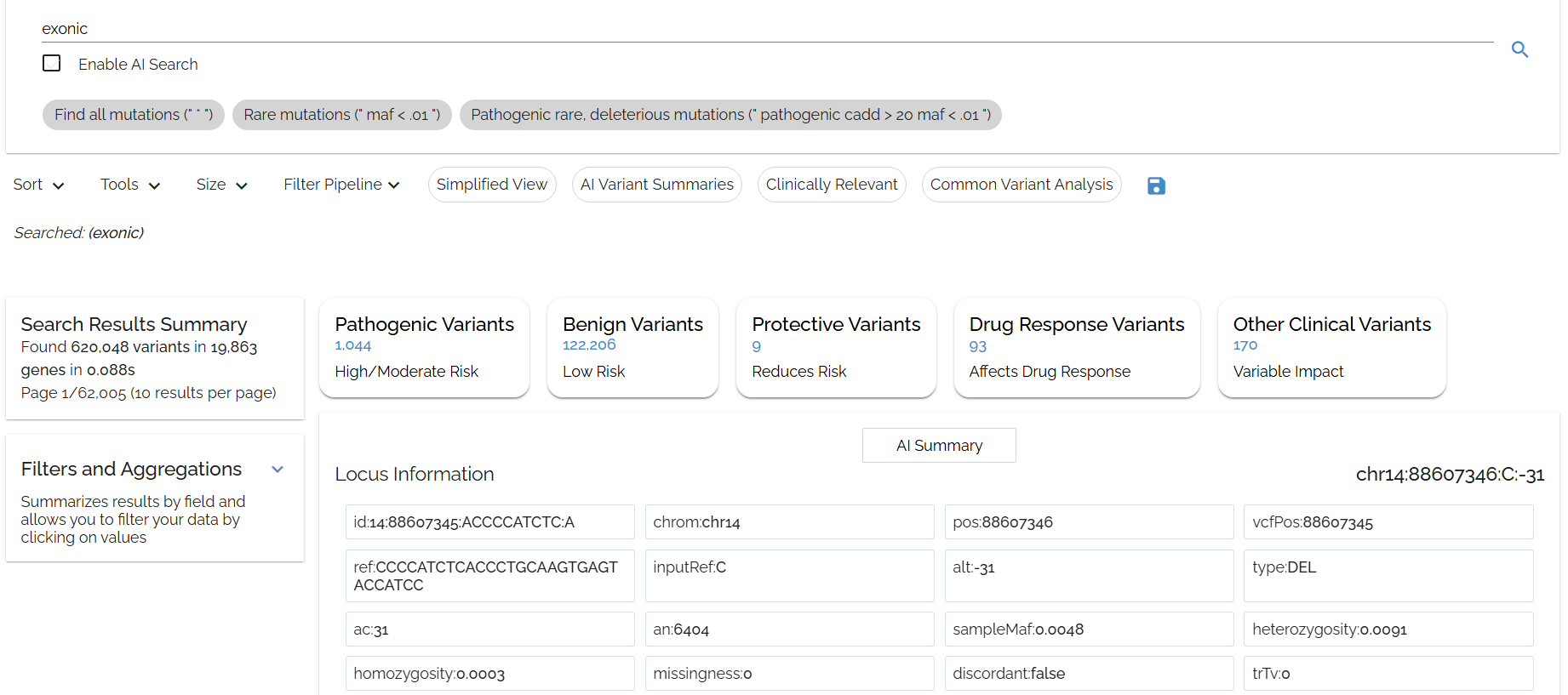
Typing in: exonic
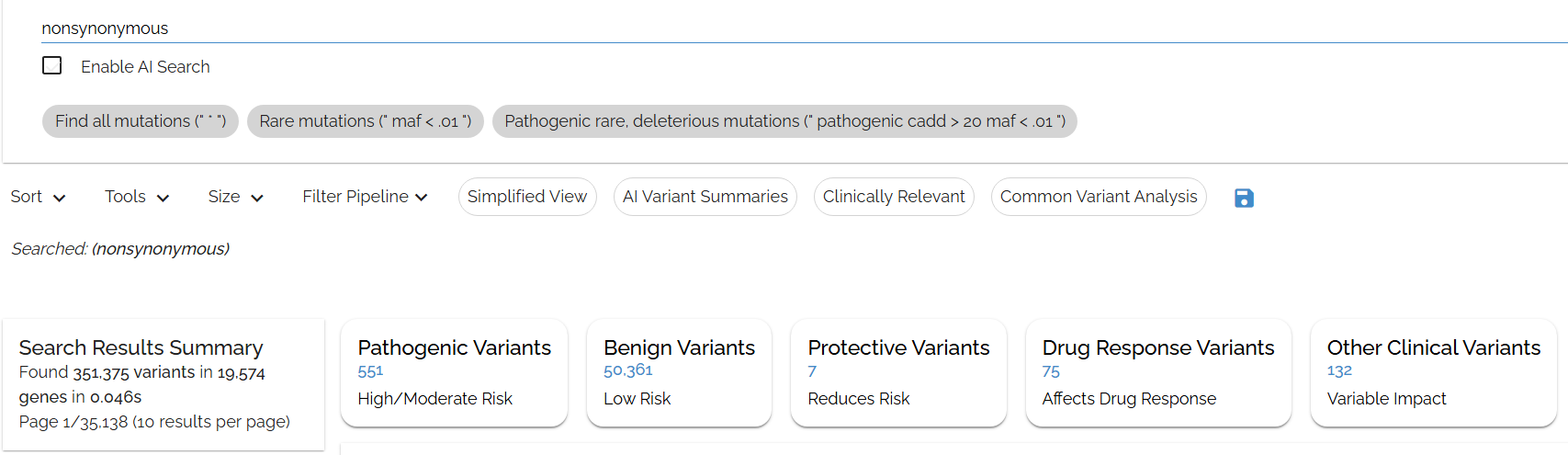
Typing in: nonsynonymous
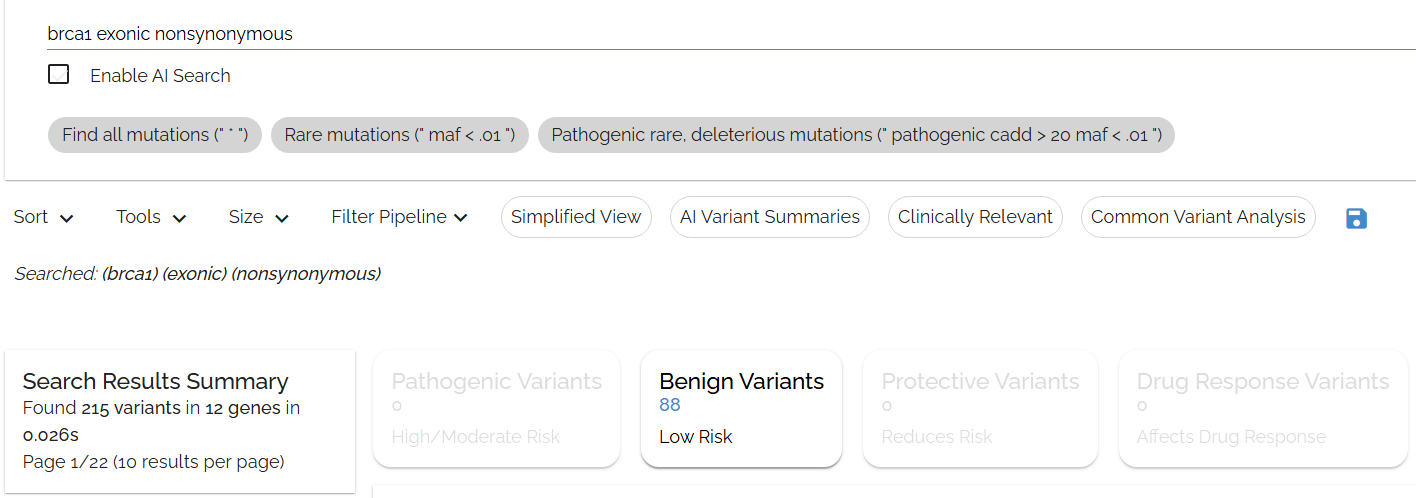
Longer queries give better results
Typing in: brca1 exonic nonsynonymous
- ▶To match, all 3 terms must be found in any field in the annotation
- ▶Note that multiple genes come up for brca1 - any fields that match this term will appear for this query, including in
refSeq.descriptionfor genes that are 'brca1-associated'. - ▶Queries can easily be refined with the filter tools.
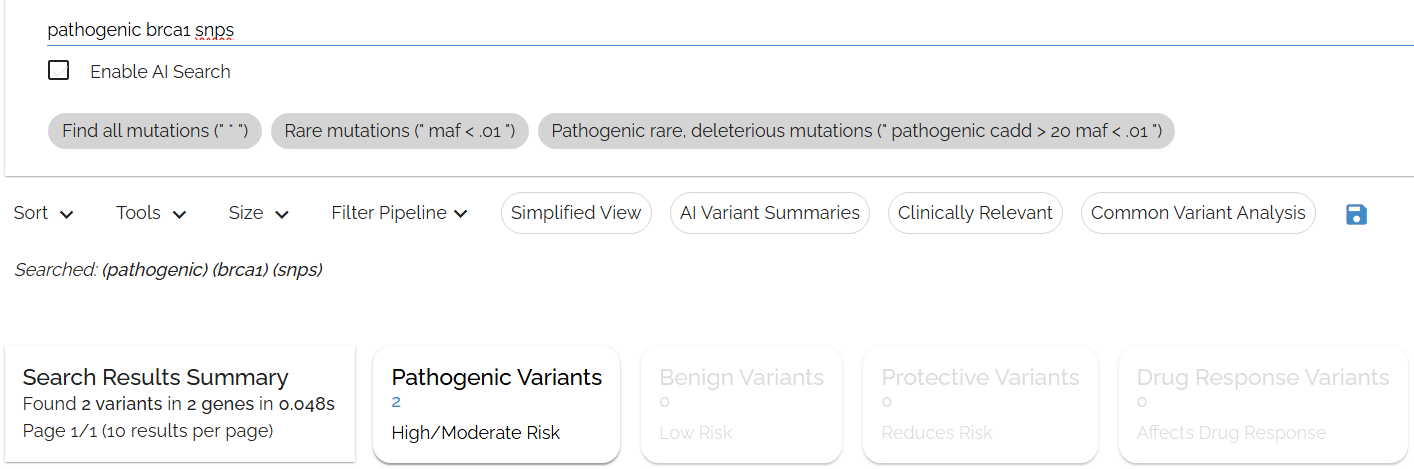
Typing in: pathogenic brca1 snps
- ▶Generally, plural and singular forms work equally well (e.g
snpvs.snps)
Multiple natural language terms can also facilitate search
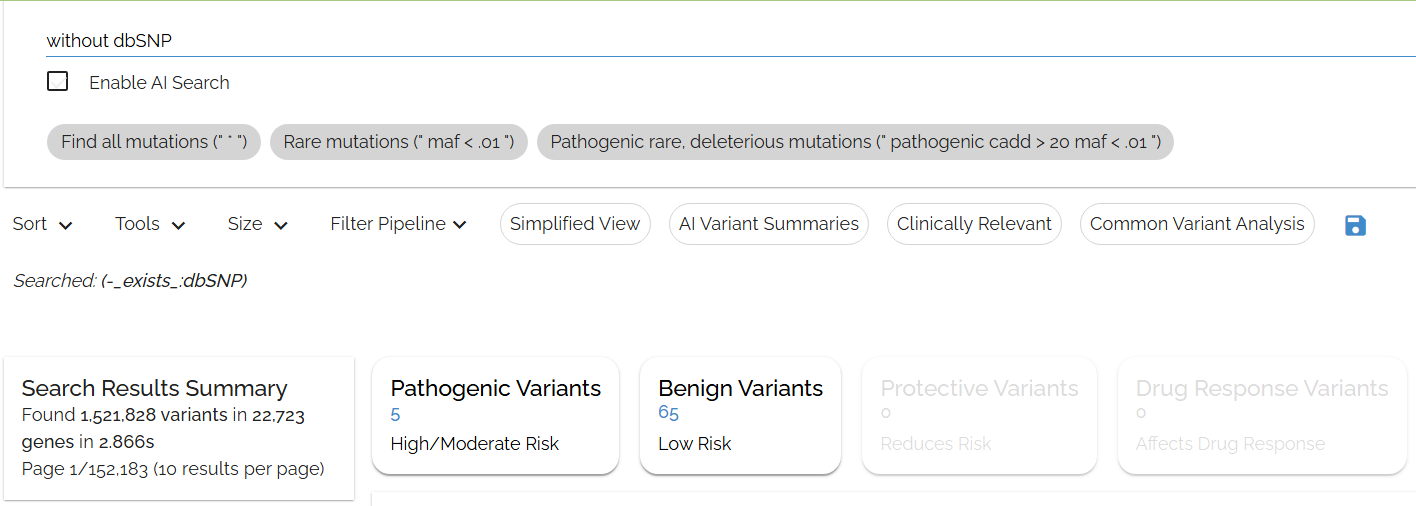
Typing in: without dbsnp
Increase specificity by using field names
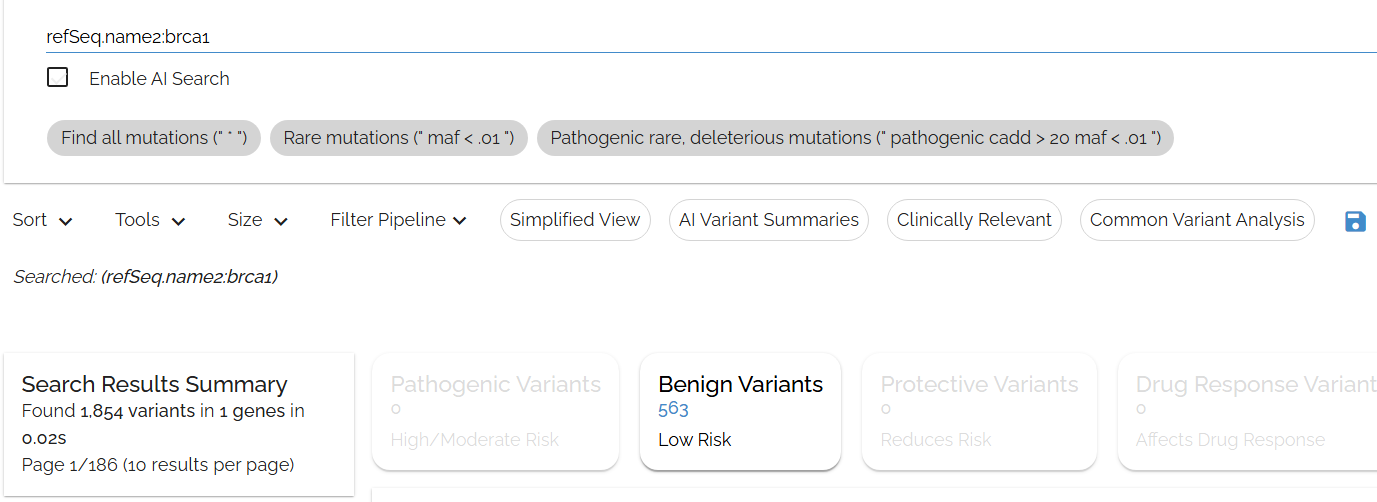
We can be more specific by searching for brca1 in the refSeq.name2 field: refSeq.name2:brca1
- ▶
name2is UCSC's field for the refSeq gene name (sometimes called a gene symbol) - ▶Any Bystro annotation field can be searched in this way
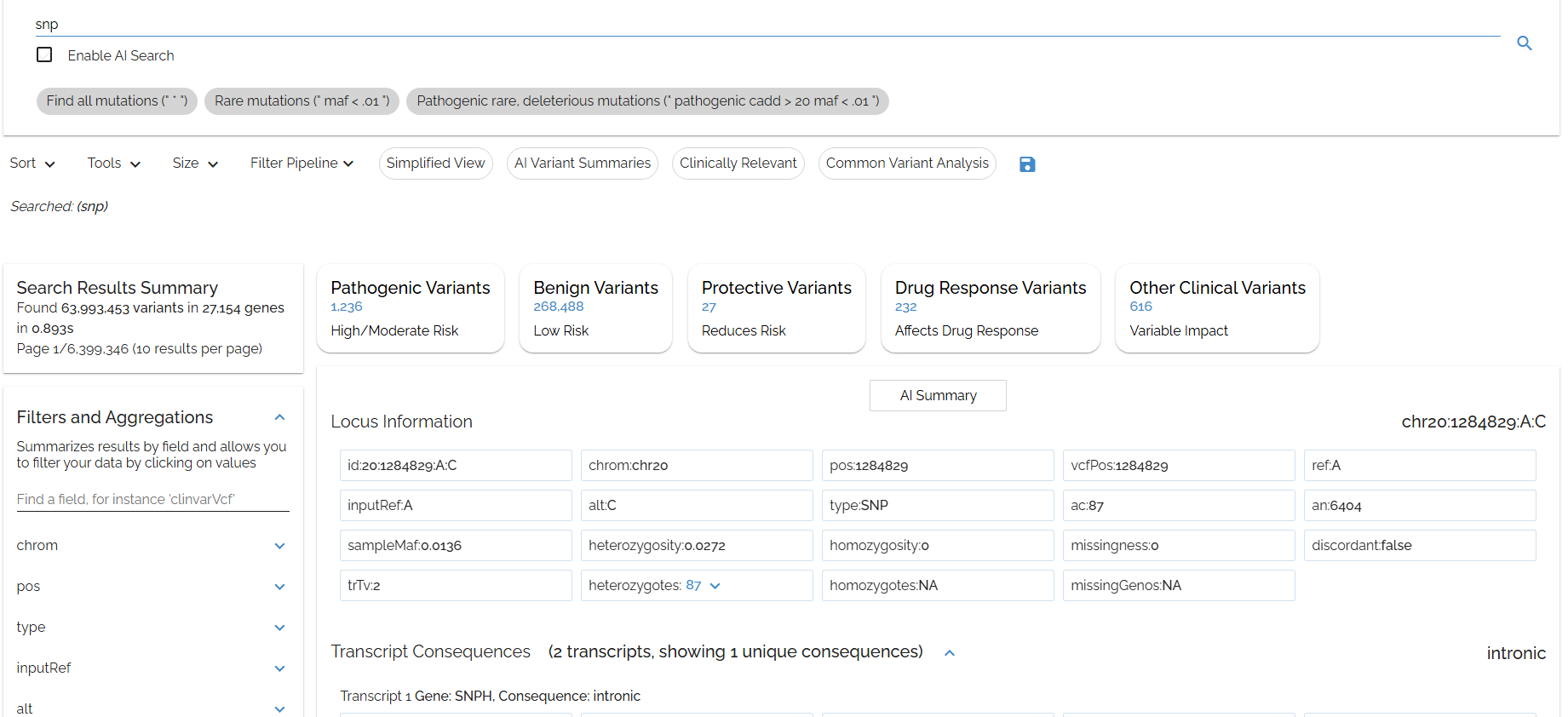
Let's try searching snp to find all single nucleotide polymorphisms
- ▶
snpmatches not only snps but also the SNPH gene
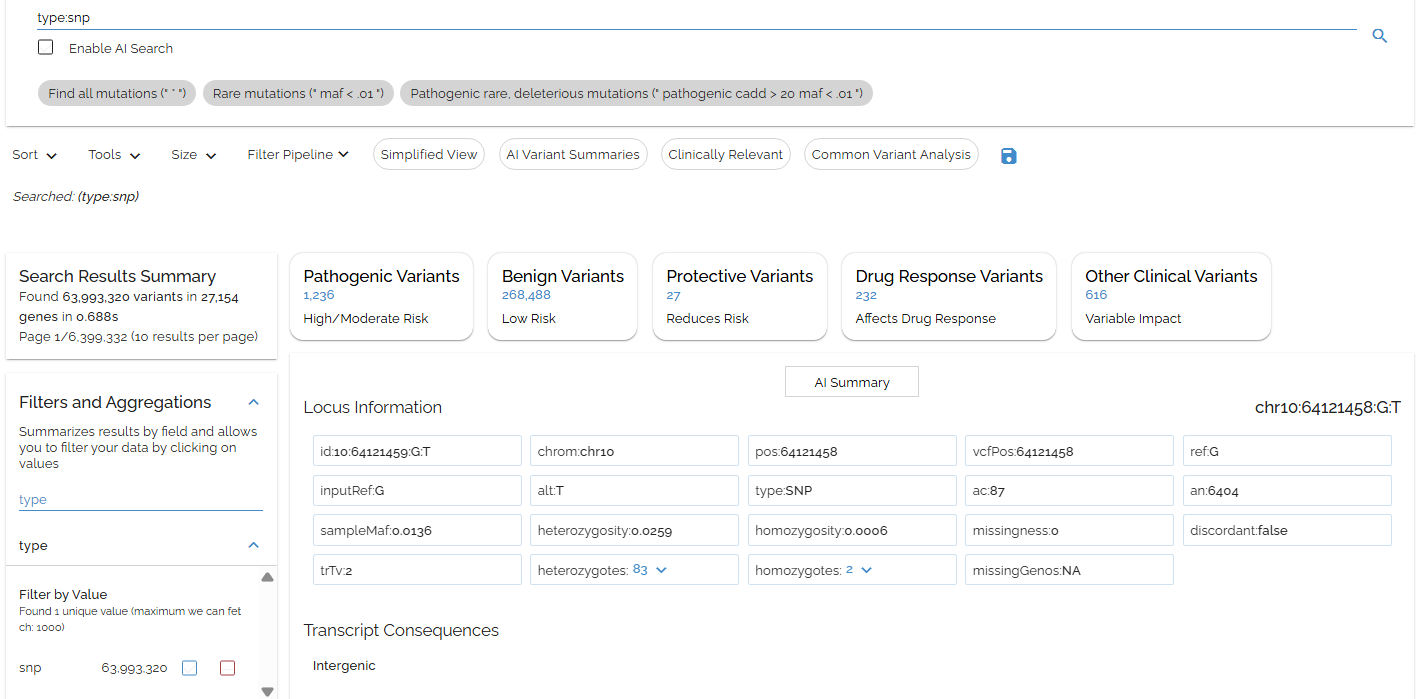
Searching the type field gives the wanted result: type:snp
- ▶The type field contains the variant call type
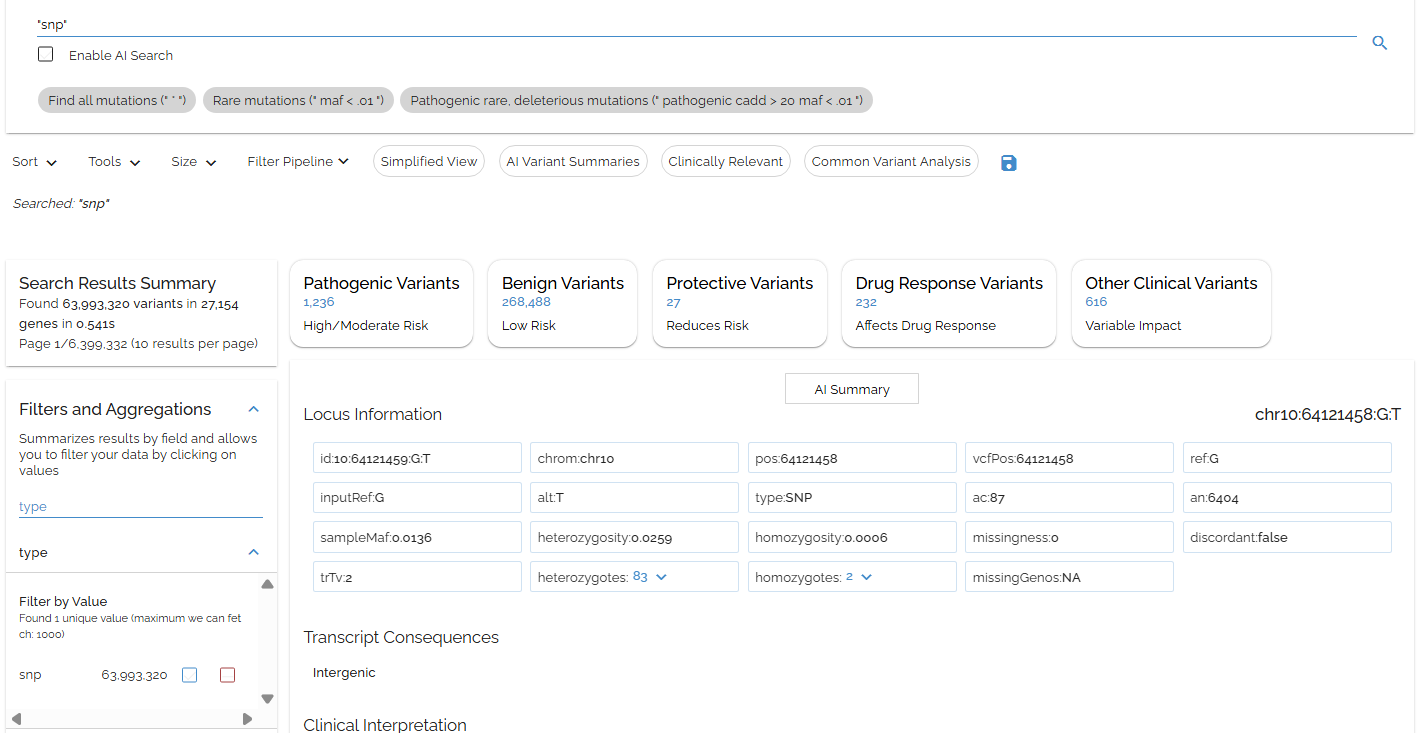
We can also solve this by searching "snp" (quotes included)
- ▶Quotes let us search phrases, which may be more specific than individual terms
- ▶(for refSeq.name2 and refSeq.nearest.name2 fields, quotes mean exact match)
Performance Note
Dataset used in examples: 1000 Genomes Project (73,452,337 variants in 27,192 genes, queries typically complete in ~0.5 seconds)