Query Allele Frequency
Filter variants by population allele frequency using gnomAD data, the largest database of human genetic variation. Identify rare, common, or population-specific variants for your research.
About gnomAD
The Genome Aggregation Database (gnomAD) is the largest collection of human genetic variation data, containing allele frequencies from diverse global populations.
Learn more about gnomAD in their official overview.
Quick Filtering with MAF
The simplest way to filter by rarity is using minor allele frequency (MAF). This searches across both gnomAD genomes and exomes datasets simultaneously for comprehensive population frequency filtering.
Common MAF Thresholds
Ultra-rare
maf < 0.001< 0.1% frequency
Rare
maf < 0.01< 1% frequency
Low frequency
maf < 0.05< 5% frequency
Example: Finding Rare Variants
Let's find rare variants (frequency < 1%) usingmaf < 0.01:
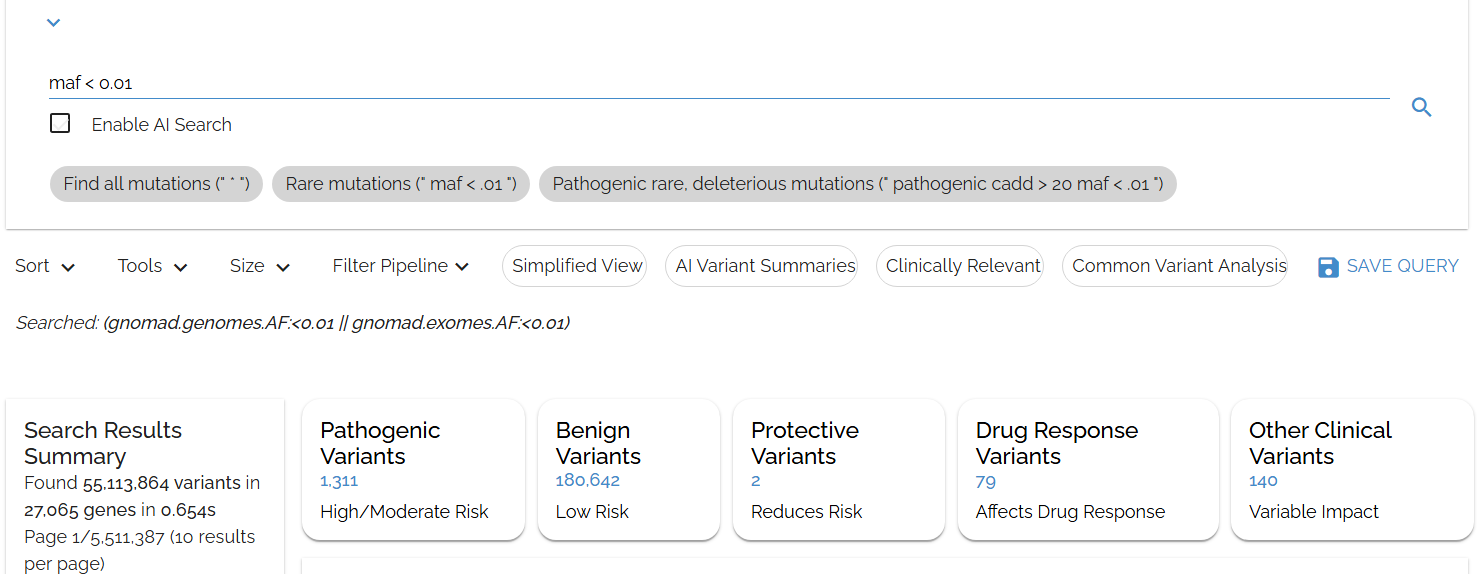
Using MAF to quickly identify rare variants across all gnomAD populations
MAF Search Benefits
Using maf as a query automatically searches both gnomAD genomes and gnomAD exomes datasets, providing comprehensive frequency filtering without needing to specify individual datasets.
Population-Specific Searches
For more precise population genetics analysis, query specific gnomAD population datasets using exact field names. This allows you to identify variants that are rare in one population but common in another.
Example: Non-Finnish European Population
Search for variants rare in the Non-Finnish European population usinggnomAD.genomes.af_nfe < 0.01:
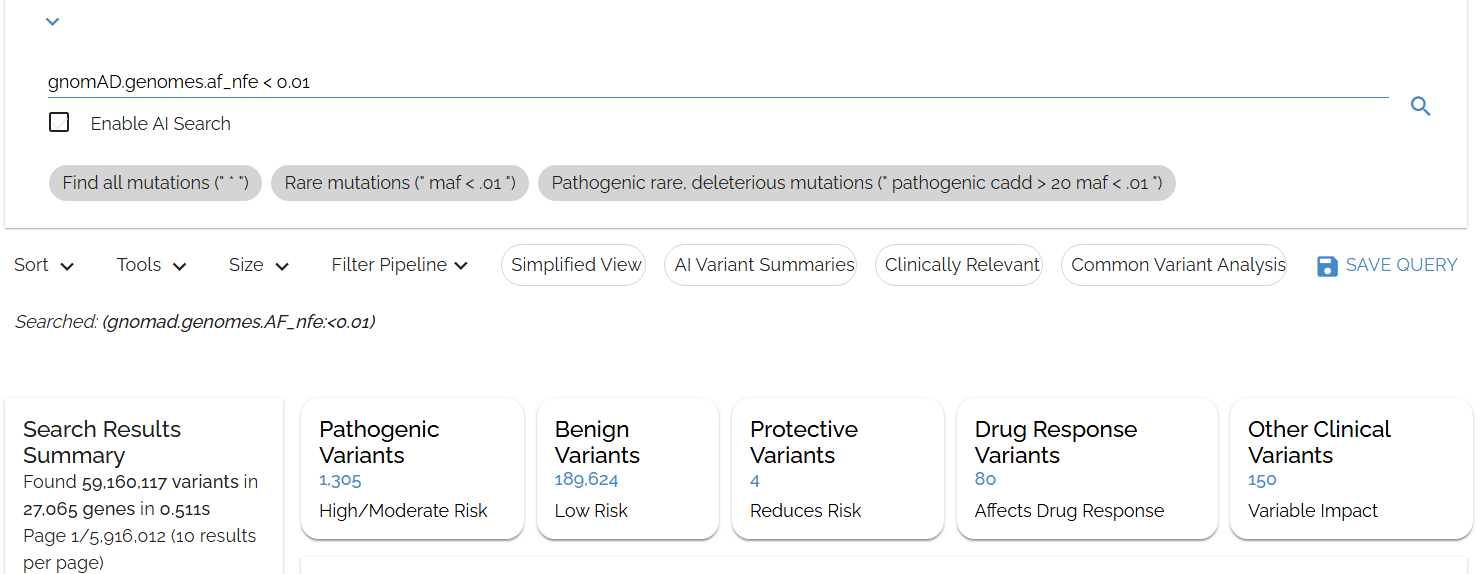
Filtering for variants rare in the Non-Finnish European population using specific gnomAD field names
Available gnomAD Populations
gnomAD Genomes Populations
- •
gnomAD.genomes.af- All populations - •
gnomAD.genomes.af_afr- African/African American - •
gnomAD.genomes.af_amr- Latino/Admixed American - •
gnomAD.genomes.af_asj- Ashkenazi Jewish - •
gnomAD.genomes.af_eas- East Asian - •
gnomAD.genomes.af_fin- Finnish - •
gnomAD.genomes.af_nfe- Non-Finnish European - •
gnomAD.genomes.af_oth- Other
gnomAD Exomes Populations
- •
gnomAD.exomes.af- All populations - •
gnomAD.exomes.af_afr- African/African American - •
gnomAD.exomes.af_amr- Latino/Admixed American - •
gnomAD.exomes.af_asj- Ashkenazi Jewish - •
gnomAD.exomes.af_eas- East Asian - •
gnomAD.exomes.af_fin- Finnish - •
gnomAD.exomes.af_nfe- Non-Finnish European - •
gnomAD.exomes.af_sas- South Asian
Complete Field Reference
Bystro reports many gnomAD fields including all population-specific frequencies. See our complete field descriptions for detailed information about all available annotation fields.
Important Considerations
Allele Frequency Reporting
Bystro reports allele frequencies relative to the specific variant allele in your dataset, not all previously observed variants at that position (which is how dbSNP reports frequencies).
gnomAD IDs
gnomAD ID shows only one rs-number per variant. Learn how this can be used to create Set IDs for SKAT analysis in our FAQ section.
Practical Applications
Rare Disease Research
Combine ultra-rare frequency filtering with high CADD scores to identify potentially pathogenic variants:
maf < 0.001 AND cadd > 20Population Genetics
Compare allele frequencies between populations to identify population-specific variants:
gnomAD.genomes.af_eas > 0.05 AND gnomAD.genomes.af_nfe < 0.01Clinical Variant Filtering
Focus on clinically relevant rare variants in coding regions:
maf < 0.01 AND refSeq.exonicAlleleFunction:nonSynonymousAllele Frequency Best Practices
• Consider your study population: Use population-specific frequencies when studying specific ethnic groups
• Account for sample size: Check allele count (AC) and allele number (AN) fields for reliability
• Combine datasets: Use both exomes and genomes data for comprehensive frequency assessment
• Validate rare variants: Always validate ultra-rare variants with additional evidence
Performance Note
Dataset used in examples: 1000 Genomes Project (73,452,337 variants in 27,192 genes, queries typically complete in ~0.5 seconds)