Query Variant Coordinates
Search for variants by genomic position using chromosome and coordinate information. Bystro supports multiple coordinate formats and range queries for flexible genomic searches.
Method 1: Field-Based Search
Search using the chrom andpos fields separately. This method gives you precise control over each component of the genomic coordinate.
Field Format Guidelines
- ▶
chromfield uses UCSC conventions (e.g., chr1, chr2, chrX, chrY) - ▶
posaccepts scientific notation (1e3), decimal (1000), or comma-separated (1,000) - ▶Syntax:
fieldname:value(e.g.,chrom:chr1 pos:2e7)
Let's search for chrom:chr19 pos:44908822, which contains one of the diagnostic Alzheimer's disease-associated APOE mutations:
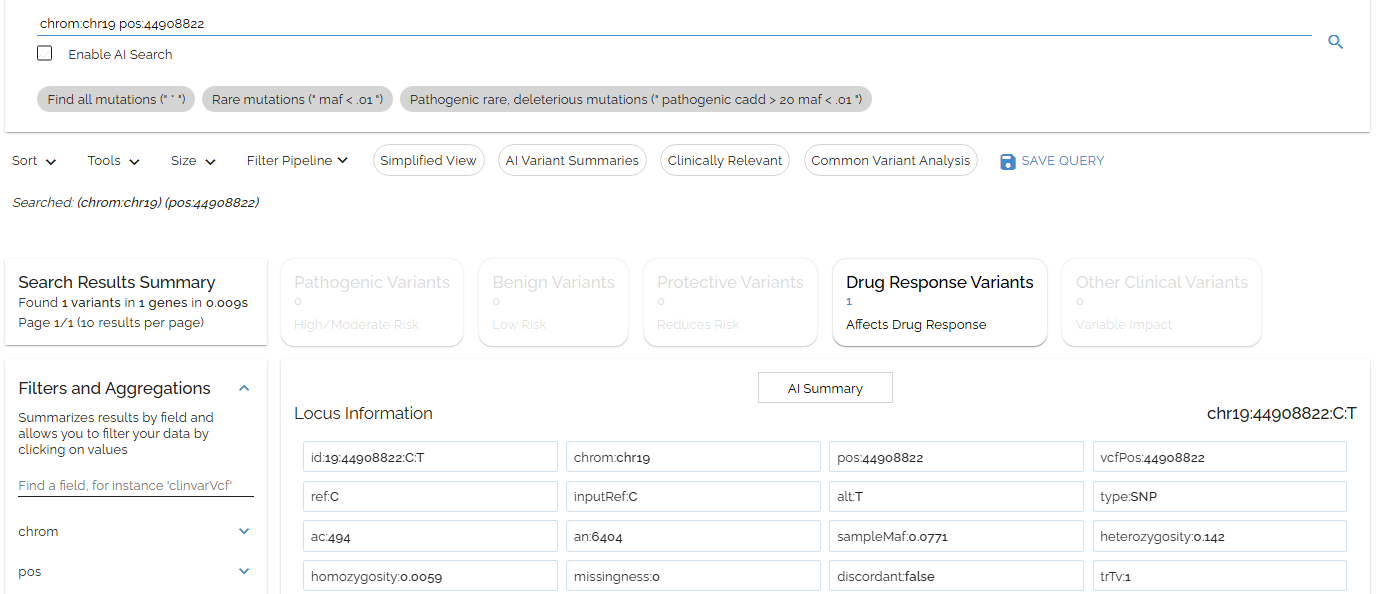
Searching for APOE variant using separate chrom and pos fields
Method 2: Coordinate Format Search
Use the familiar chrN:position format for quick coordinate searches. This automatically searches both the chrom and pos fields.
Accepted Formats
- •
chr17:41,215,920(with commas) - •
chr17:41215920(without commas) - •
chr17:4.12e7(scientific notation)
Benefits
- • Familiar genomics format
- • Faster for single position queries
- • Compatible with genome browsers
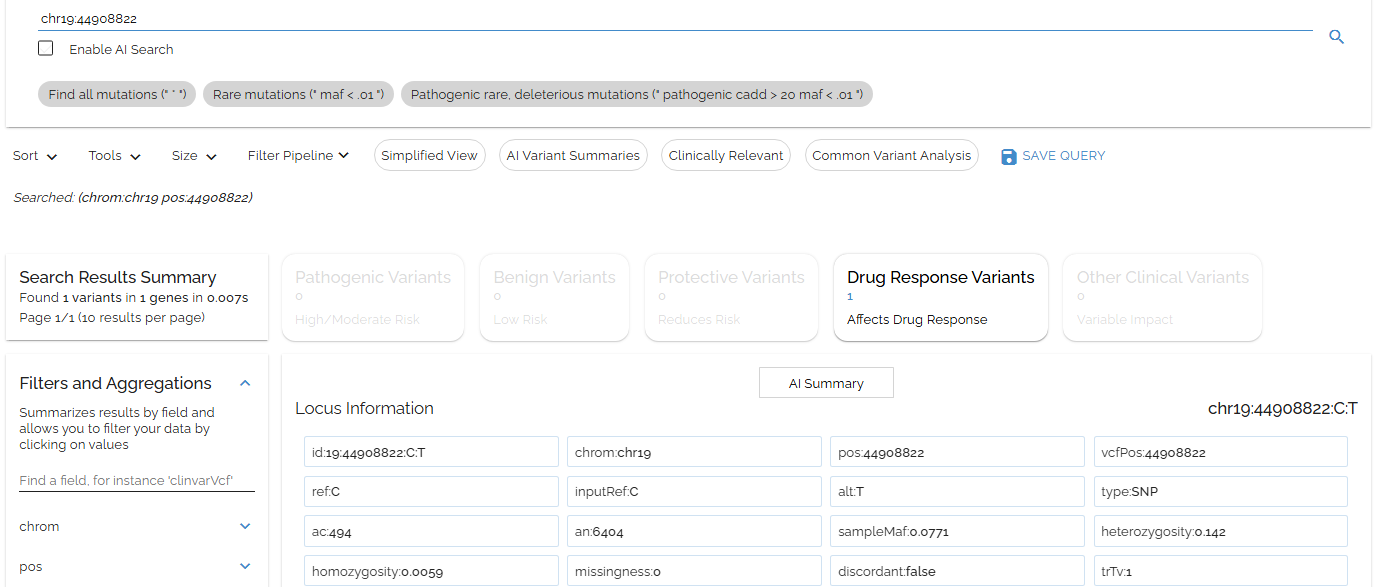
Same APOE search using the convenient chr19:position format
Position Range Queries
Search genomic regions using range syntax to find all variants within a specific interval. This is particularly useful for analyzing genes, regulatory regions, or chromosomal segments.
Range Query Syntax
Recommended Format:
- •
pos:[start TO end] - • Brackets are optional but safer for complex queries
- •
TOkeyword is explicit and clear
Alternative Formats:
- •
pos1 - pos2(dash separator) - •
pos1 ... pos2(ellipsis separator)
Range Query Caution
Be careful with the dash (-) operator as it has multiple meanings:
- • Numeric sign:
-100(negative number) - • Exclusion operator:
-termToExclude(exclude results containing this term) - • Range separator:
pos1 - pos2(position range)
Use TO syntax for clarity in complex queries.
Example: 3q29 Microdeletion Region
Let's search the 1.6Mb 3q29 region associated with microdeletion syndrome using chr3:195.7e6 - 197.4e6:
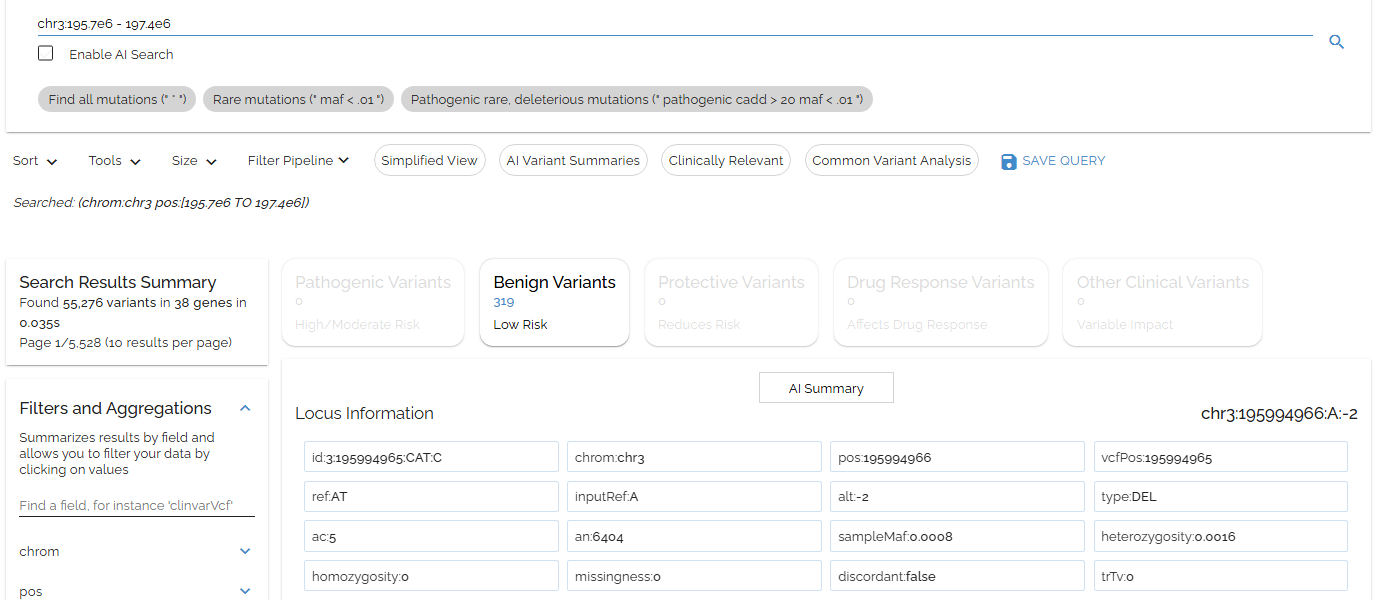
Searching the 3q29 microdeletion region (chr3:195.7M-197.4M) to find all variants in this clinically relevant interval
Practical Applications
Single Variant Lookup
Find specific known variants by exact coordinates
chr17:41215920Gene Region Analysis
Search entire genes or regulatory regions
chrom:chr17 pos:[41.1e6 TO 41.3e6]Chromosomal Segments
Analyze large genomic regions or cytogenetic bands
chr3:195e6 - 200e6Coordinate Search Tips
• Combine with filters: Use coordinate searches with functional annotations for targeted analysis
• Scientific notation: Use 1e6 for millions, 1e3 for thousands to avoid typing errors
• Check genome build: Ensure your coordinates match the reference genome used in your dataset
• Range queries: Consider using gene names for known genes rather than manual coordinate ranges
Performance Note
Dataset used in examples: 1000 Genomes Project (73,452,337 variants in 27,192 genes, queries typically complete in ~0.5 seconds)