Create Custom Search Shortcuts
Create powerful search shortcuts that let you query multiple genes, pathways, sample groups, or complex conditions with a single word. Transform repetitive, complex searches into simple, reusable commands tailored to your research.
Why Custom Shortcuts Are Powerful
Instead of repeatedly typing complex queries likeBRCA1 OR BRCA2 OR PALB2 OR CHEK2, create a shortcut called breastCancerGenesand just search for that.
Perfect for gene panels, sample groups, pathway analysis, and case-control studies.
How to Create Search Shortcuts
Start with any search
Begin by searching for anything (you can use * to see all results) to access the tools menu.
Open the synonym creator
Click "Custom Synonyms": below the search bar
Define your shortcut
In the popup, enter a memorable label (your shortcut name) and list the values you want it to represent, one per line.
Save and search
Click Save, then use your new shortcut label in any search. Bystro will automatically expand it to search all your defined values.
How Shortcuts Work
Behind the Scenes
What You Type
breastCancerGenesWhat Bystro Searches
BRCA1 OR BRCA2 OR PALB2 OR CHEK2Your shortcut label is automatically replaced with all your synonym values joined byOR operators.
Powerful Examples
Gene Panel Shortcuts
Create shortcuts for gene panels you frequently analyze. Instead of typing multiple gene names every time, define them once and reuse:
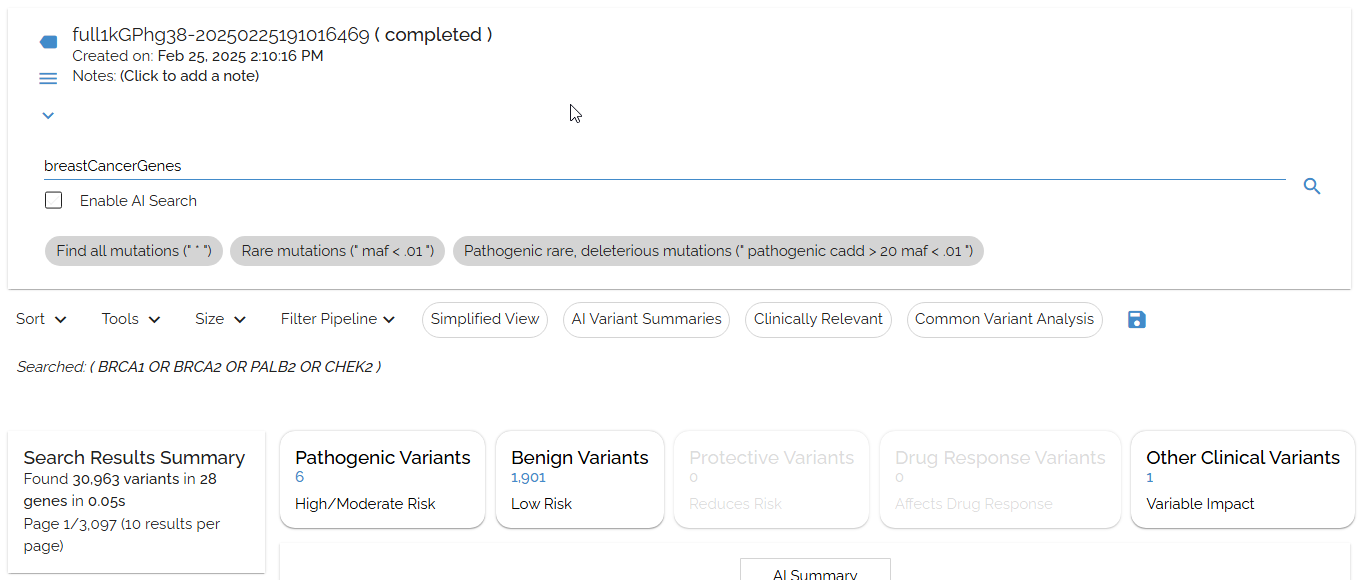
Creating a 'breastCancerGenes' shortcut to search BRCA1, BRCA2, PALB2, and CHEK2 simultaneously
Field-Specific Shortcuts
Combine your shortcuts with specific annotation fields for precise searches. Use refSeq.name2:breastCancerGenes to search only in gene name fields:
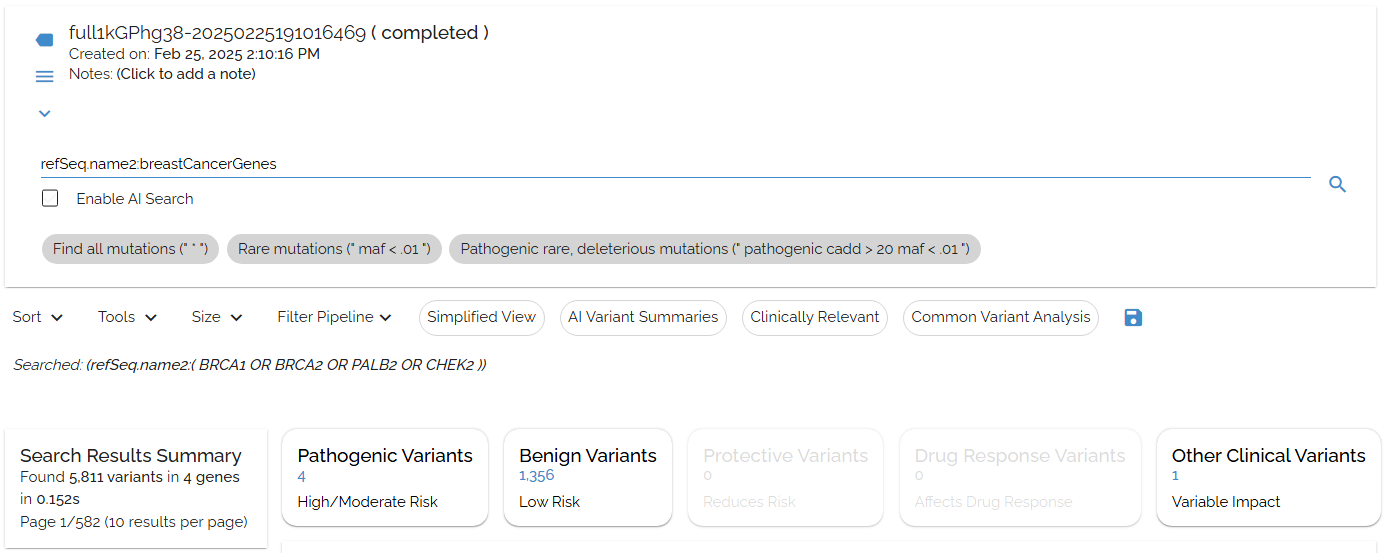
Using gene panel shortcuts with specific annotation fields for targeted searches
Complex Query Shortcuts
Synonym values can be complete queries, not just simple terms. Create shortcuts for complex filtering conditions you use repeatedly.
Advanced Shortcut Techniques
Complex Queries as Values
Each synonym value can be a full query with operators and conditions. This lets you create shortcuts for sophisticated filtering criteria you use repeatedly:
Shortcut: rarePathogenic
gnomad.genomes.af < 0.001clinvar.clinicalSignificance:Pathogeniccadd.phred > 20Exact Matching with Quotes
Use quotes to define phrase boundaries and create more precise matches. This prevents partial matches and ensures you get exactly the genes you want:
Shortcut: alzheimerGenes
"APOE""PSEN1""PSEN2""APP"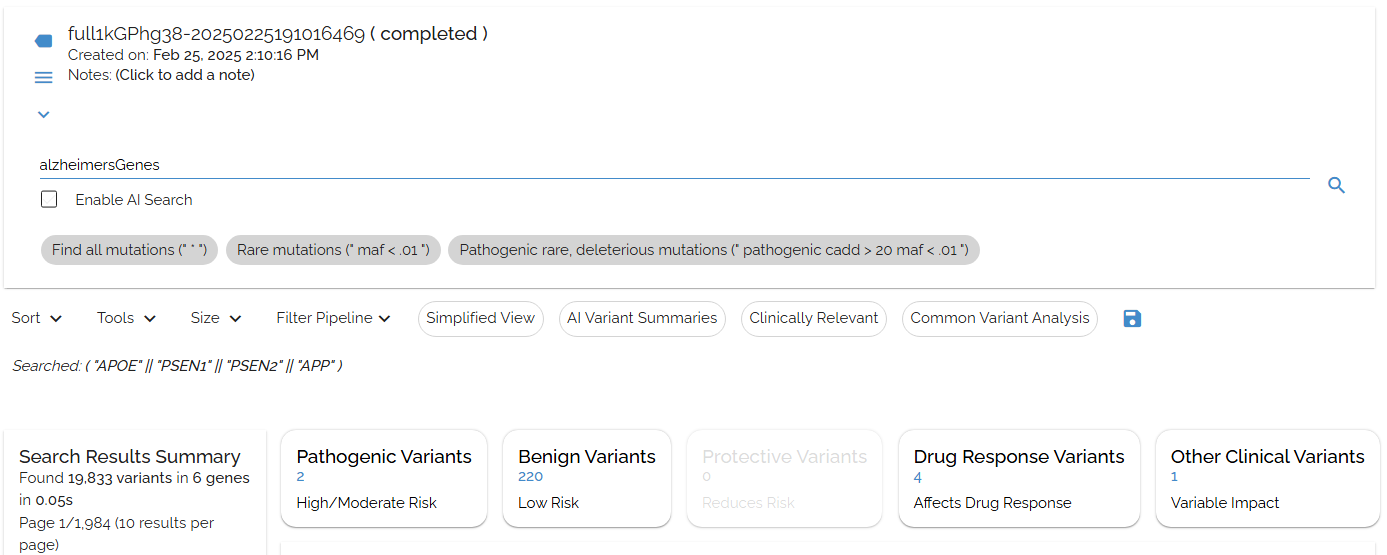
Creating precise gene shortcuts using quotes for exact matching with Alzheimer's disease genes
Case-Control and Family Studies
Custom shortcuts excel at organizing sample groups for comparative analysis. Combine with boolean operators for powerful study designs:
Sample Group Examples
Case-Control Studies
- • Create
casesshortcut with affected sample IDs - • Create
controlsshortcut with unaffected sample IDs - • Search
cases -controlsfor case-specific variants
Family Studies
- • Create
probandsshortcut with affected children - • Create
parentsshortcut with parent sample IDs - • Search
probands -parentsfor de novo variants
Boolean Operators with Shortcuts
Use the NOT operator (-) to find mutually exclusive sets:
- •
cases -controls→ variants only in cases - •
children -parents→ de novo variants - •
tumor -normal→ somatic mutations
Creative Applications
Pathway Analysis
Group genes by biological pathways for functional enrichment studies.
DNArepairTissue-Specific Panels
Create organ or tissue-specific gene lists for targeted analysis.
cardiacGenesCohort Subgroups
Organize samples by demographics, treatment response, or phenotype.
respondersShortcut Management Tips
• Use descriptive names: Choose memorable labels that clearly describe the content
• One value per line: Each synonym value should be on its own line in the creator
• Test your shortcuts: Verify they work as expected before using in important analyses
• Document complex shortcuts: Keep notes about what each shortcut contains for future reference
Performance Note
Dataset used in examples: 1000 Genomes Project (73,452,337 variants in 27,192 genes, queries typically complete in ~0.5 seconds)