Incomplete/Failed Annotations
Monitor your annotation progress and troubleshoot issues when annotations don't complete successfully. Bystro provides dedicated pages to track incomplete jobs and diagnose failures.
📍 Where to Find Your Annotations
⏳ In Progress: Check the Incomplete page for annotations currently processing
❌ Failed: Visit the Failed page for annotations that encountered errors
Tracking Annotation Progress
The Incomplete page shows real-time progress for your running annotations, allowing you to monitor large jobs and estimate completion times.
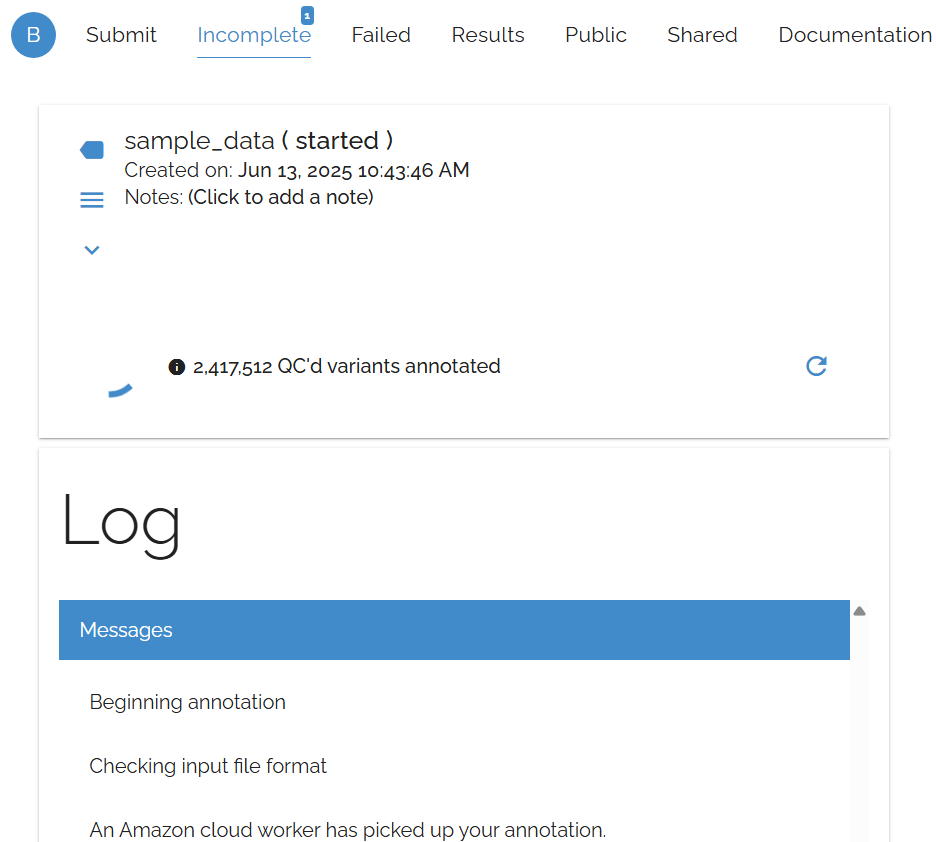
Progress tracking interface showing annotation status and completion estimates
⏳ Incomplete Annotations
Currently processing jobs with progress indicators and estimated completion times. Large datasets may take several hours to complete.
✅ Completed Annotations
Successful annotations automatically move to the Results page where you can download and analyze your annotated data.
Understanding Failed Annotations
Failed annotations are rare but can occur due to file format issues, incompatible genome builds, or other technical problems. When they do happen, the system provides specific error information to help you resolve the issue.
Common Failure Causes
- ▶Wrong genome build: VCF coordinates do not match selected reference genome
- ▶File format issues: Malformed VCF headers or invalid file structure
- ▶Incompatible file types: Files that appear to upload but contain unsupported data
- ▶Corrupted uploads: Files damaged during transfer or compression
Troubleshooting Steps
Step 1: Check the Error Message
Review the specific error type displayed on the Failed page. This usually indicates the root cause of the problem.
Step 2: Verify Genome Build
Double-check that you selected the correct assembly/genome build (e.g., hg19, hg38, GRCh37, GRCh38) that matches your VCF file coordinates.
Step 3: Validate File Format
Ensure your VCF file has proper headers and follows standard formatting. Test with a smaller subset of your data if possible.
Step 4: Resubmit
After addressing potential issues, try resubmitting your file. Most problems can be resolved by correcting the genome build or file format.
Getting Additional Help
If you continue to experience issues after following the troubleshooting steps:
- ▶Check our FAQ:Visit theFAQ pagefor common issues and solutions
- ▶Contact support: Reach out with specific error messages and file details
- ▶Community forum: Search for similar issues or ask the community
💡 Prevention Tips
- ▶Test with small files first: Validate your workflow with a subset of data
- ▶Use standard tools: Generate VCFs with well-established pipelines
- ▶Double-check genome builds: Verify reference genome before submission
- ▶Monitor progress: Check the Incomplete page for long-running jobs